Nanopore Analysis of DNA
In collaboration with the Burrows group (University of Utah) we have a continuing interest in the use of biological nanopores to interrogate DNA structure. Much of our current work focuses on the use of the protein pore alpha-hemolysin (αHL) particularly for the identification of DNA damage. DNA damage is a significant issue in human cells, to take just one example de-purination of adenine occurs approximately 18,000 times in each cell per day. Unrepaired damage leads to mutations that in turn are responsible for the formation of cancers.
Under an applied potential, negatively charged DNA can be driven into the protein pore. Single-stranded DNA translocates through the pore with a characteristic translocation time, and attenuates the measured current as it moves through the pore. Both the translocation time and the current attenuation are characteristic of the DNA composition. We have found that the introduction of bulky damage sites in single-stranded DNA (e.g. Benoz[a]pyrene) will perturb translocation time and increase ion flux attenuation.
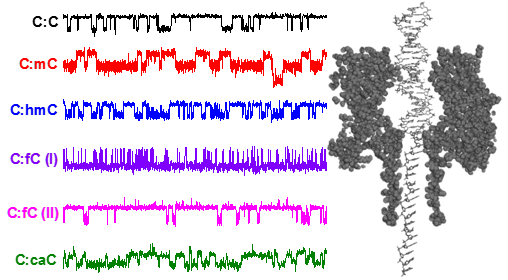
While ssDNA can pass through the smallest constriction in αHL (1.4 nm), dsDNA, which has a nominal diameter of 2.0 nm, cannot. However, it is possible to capture dsDNA inside the α-HL vestibule, and with an appropriate voltage (> 100 mV) the dsDNA duplex will unzip into its constituent single-stranded components. The residence time prior to unzipping is dependent on the composition of the duplex. We have recently discovered that structural modifications in dsDNA can be detected from the magnitude to which ion flow is attenuated during dsDNA residence inside the protein pore. The precise change in current is dependent on the position of the missing base in the sequence relative to the latch constriction of the α-HL protein pore, a newly discovered sensing zone in DNA (see figure below). We are currently exploring uses of this sensing zone for identifying and discriminating damage sites in dsDNA.
Select Publications:
γ-Hemolysin Nanopore Is Sensitive to Guanine-to-Inosine Substitutions in Double-Stranded
DNA at the Single-Molecule Level
Cherie S. Tan, Aaron M. Fleming , Hang Ren , Cynthia J. Burrows, and Henry S. White
J. Am. Chem. Soc., 2018, 140 (43), pp 14224–14234.
Single-Molecule Titration in a Protein Nanoreactor Reveals the Protonation/Deprotonation
Mechanism of a C:C Mismatch in DNA
Hang Ren, Cameron G. Cheyne, Aaron M. Fleming, Cythia J. Burrows, and Henry S. White
J. Am. Chem. Soc., 2018, 140, 15, 5153-5160
Nanopore Analysis of the 5‑Guanidinohydantoin to Iminoallantoin Isomerization in Duplex
DNA
T. Zeng, A. M. Fleming, Y. Ding, H. Ren, H. S. White, C. J. Burrows
J. Org. Chem., 2018, 83 (7), pp 3973–3978
Dynamics of a DNA Mismatch Site Held in Confinement Discriminate Epigenetic Modifications
of Cytosine
R. P. Johnson, A. M. Fleming, R. T. Perera, C. J. Burrows, and H. S. White
J. Am. Chem. Soc., 2017, 139 (7), 2750–2756.
Interrogation of Base Pairing of the Spiroiminodihydantoin Diastereomers Using the
α-Hemolysin Latch
T. Zeng, A. M. Fleming, Y. Ding, H. S. White, and C. J. Burrows
Biochemistry, 2017, 56 (11), 1596–1603.
Energetics of base flipping at a DNA mismatch site confined at the latch constriction
of α-hemolysin
R. P. Johnson, R. T. Perera, A. M. Fleming, C. J. Burrows and H. S. White
Faraday Discuss., 2016, 193, 471-485.
Detection of benzo [a] pyrene-guanine adducts in single-stranded DNA using the α-hemolysin
nanopore
R. T. Perera, A. M. Fleming, R. P. Johnson, C. J. Burrows, and H. S. White
Nanotechnology, 2015, 26(7), 074002.
Temperature and electrolyte optimization of the a-hemolysin latch sensing zone for
detection of base modification in double-stranded DNA
R. P. Johnson, A. M. Fleming, Q. Jin, C. J. Burrows, and H. S. White
Biophys. J. , 2014, 107(4), 924-931.
Effect of an Electrolyte Cation on Detecting DNA Damage with the Latch Constriction
of α-Hemolysin
R. P. Johnson, A. M. Fleming, C. J. Burrows, and H. S White
J. Phys. Chem. Lett., 2014, 5(21), 3781-3786.
Base-Excision Repair Activity of Uracil-DNA Glycosylase Monitored Using the Latch
Zone of α-Hemolysin
Q. Jin, A. M. Fleming, R. P. Johnson, Y. Ding, C. J. Burrows, and H. S. White
J. Am. Chem. Soc., 2013, 135(51), 19347-19353.
Base-Excision Repair Activity of Uracil-DNA Glycosylase Monitored Using the Latch
Zone of α-Hemolysin
Q. Jin, A. M. Fleming, R. P. Johnson, Y. Ding, C. J. Burrows, and H. S. White
J. Am. Chem. Soc., 2013, 135(51), 19347-19353.