Research Interests
Research efforts in the Flynn group emphasizes biophysical characterization of biologically active macromolecules using multidimensional/multinuclear solution NMR methods. Our approach emphasizes development of in vitro models of the cellular environment, and explores the influence of crowded environments on the behavior of proteins and small RNA oligonucleotide model systems.
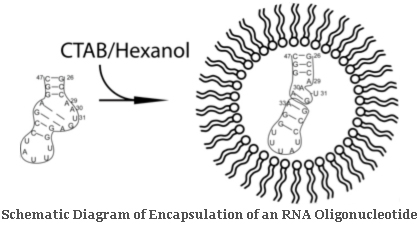
Encapsulation of MacromoleculesReverse Micelle Model
 Proteins, nucleic acid oligomers, and other biologically important macromolecules
may be encapsulated within a surfactant shell (i.e., dioctyl sulfosuccinate) and transported
into non-native solvent systems (i.e., short-chain n-alkanes). This novel approach
opens up new opportunities for solution NMR studies of larger macromolecules, as well
as providing a unique and powerful new approach to studies of the environment on the
physical behavior of macromolecules. Current group effort are focused on the development
of encapsulation as model of the crowded intracellular environment.
Proteins, nucleic acid oligomers, and other biologically important macromolecules
may be encapsulated within a surfactant shell (i.e., dioctyl sulfosuccinate) and transported
into non-native solvent systems (i.e., short-chain n-alkanes). This novel approach
opens up new opportunities for solution NMR studies of larger macromolecules, as well
as providing a unique and powerful new approach to studies of the environment on the
physical behavior of macromolecules. Current group effort are focused on the development
of encapsulation as model of the crowded intracellular environment.
Macromolecular Structure and DynamicsSolution Structure of 15.5K Protein
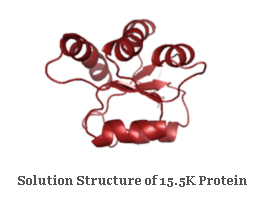 All biological function derives from the structure and dynamics of macromolecules.
Structure has long been used as the primary lead in efforts to understand the molecular
basis of function, and remains fundamentally important. The importance of internal
dynamics in proteins as a factor in characterizing function has become increasingly
apparent, and solution NMR methods are particularly well-suited to evaluate such effects.
Studies of backbone dynamics in proteins indicate that the main-chain atoms are generally
highly and homogeneously ordered whereas studies of side-chain motion suggest a more
heterogeneous picture. We are particularly interested in probing dynamics at the interfaces
between components of protein-protein and protein-RNA complexes, across a wide range
of time scales (from microseconds to hours) to characterize the full range of motion
that influence the interaction. New efforts target characterization of dynamics in
the RNA oligos, which promises to generate novel insights into the physical nature
of protein-RNA recognition events.
All biological function derives from the structure and dynamics of macromolecules.
Structure has long been used as the primary lead in efforts to understand the molecular
basis of function, and remains fundamentally important. The importance of internal
dynamics in proteins as a factor in characterizing function has become increasingly
apparent, and solution NMR methods are particularly well-suited to evaluate such effects.
Studies of backbone dynamics in proteins indicate that the main-chain atoms are generally
highly and homogeneously ordered whereas studies of side-chain motion suggest a more
heterogeneous picture. We are particularly interested in probing dynamics at the interfaces
between components of protein-protein and protein-RNA complexes, across a wide range
of time scales (from microseconds to hours) to characterize the full range of motion
that influence the interaction. New efforts target characterization of dynamics in
the RNA oligos, which promises to generate novel insights into the physical nature
of protein-RNA recognition events.
